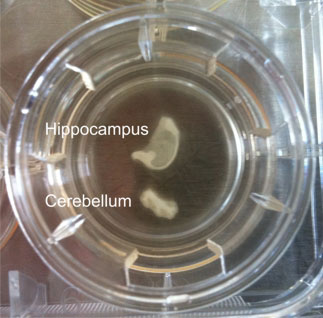
Figure 1: Hippocampal and cerebellar brain slice culture from a bovine. Anatomical architecture is maintained and at the edges cells grow out.

Anna Oevermann
Neuropathology, Division of Neurological Science
Vetsuisse Faculty, University of Bern, Switzerland
Keywords: live stock; brain; infectious diseases; veterinary disease; cell cultures: organ-specific; reduction; replacement
Duration: 3 years Project Completion: 2013
Background and Aim
Infectious disorders of the central nervous system (CNS) in livestock may have severe economic and public-health implications and are therefore of major concern. This was demonstrated dramatically in the mid 1990s, when it became evident during the upsurge of bovine spongiform encephalopathy (BSE) that the disease was transmissible from cattle to humans. Recently, atypical variants of transmissible spongiform encephalopathies (TSEs) have been detected in ruminants. As yet, it is not known whether they manifest inter-species cross-over potential. Listeriosis, which is caused by Listeria (L.) monocytogenes(LM), is another infectious and zoonotic CNS-disease of likewise high impact on livestock and humans.
Despite intensive research activity in the fields of TSEs and listeriosis during the past decades, very few in-vitro sytems are available for the modelling of their neuropathogenesis and host-pathogen interactions and for strain-typing. Studies depend largely on bioassays either in laboratory rodents or in the natural ruminant host, since these best reflect the intricate pathogenesis of CNS-infections. Given the highly invasive inoculation routes and the severity of the resulting disease, such experiments raise fundamental ethical concerns, and are awarded the highest severity grade (Schweregrad 3). In the case of TSEs, experiments may be protracted over several years until the time of sacrifice. Moreover, it is often unclear to what extent the results of rodent models can be extrapolated to the situation in the natural host. On the other hand, pertinent cell models of ruminant neuroinfectious diseases do not exist. Hence, our aim was to develop an ethically-sustainable, host-specific, organotypic brain-slice culture using material derived from regularly-slaughtered ruminants as an in-vitro system for the identification and investigation of neuroinfectious diseases, here exemplified for prion diseases and listeric encephalitis.
Method and Results
The hippocampus, cerebellum and brainstem were collected from sheep, goats and cattle at a slaugtherhouse, immediately after slaughter. Using a vibratome, 350-µm-thick brain-slices were cut and plated on membrane inserts in 6-well plates (Figure 1; for methodological details, please see Guldimann et al., 2012). We assessed the viability of the brain-slices by determining the difference between the number of dead cells and the total number of cells present in a slice (Figure 2, a and b). The presence of brain-cell types (neurons, microglia, astrocytes, oligodendrocytes) was assessed by double-immunofluorescence. We demonstrated that bovine brain-slices can be maintained in culture for up to forty-nine days and that all endogenous brain-cell populations, including neurons (Figure 3), are present. Because the viability of the brainstem and of the cortical slices was low, experiments with these regions were discontinued. The viability results of the hippocampal slices were consistently better than those of the cerebellar ones. Brain-slices that were derived from slaughtered sheep and goats were significantly less viable than were those stemming from cattle. Owing to the disintegration of the tissue, they could not be maintained for extended periods of time in culture. For these reasons, we used only bovine hippocampal slices for the infection assays with LM. To allow time for the explantation trauma to subside, the slices were incubated for seven days prior to the performance of the infection assays. We demonstrated that this in-vitro system is susceptible to LM-infection and that it replicates features of natural rhombencephalitis in ruminants (Figure 5; Guldimann et al., 2012).
To investigate whether specifically neurovirulent LM-strains can be identified and distinguished from those of other sources in the brain-slice model, infection assays with forty-seven LM-strains from different sources (rhombencephalitis, abortion, gastroenteritis, environment, food, human infections) and genetic complexes were performed on cultured bovine hippocampal slices (Guldimann et al., in preparation). To measure bacterial replication in the brain-slices, CFU’s were determined 48 hours post-infection. To evaluate the extent of bacterial spreading, the slices were fixed in paraformaldehyde, and immunofluorescence for LM was then performed. The total size of all bacterial foci within a slice was measured and expressed in µm2. All LM-strains but one were capable of infecting and of replicating in bovine hippocampal slices. Although the infection assays in brain-slice cultures cannot clearly distinguish between encephalitogenic, non-encephalitogenic and environmental LM-strains, they nevertheless reveal inter-strain differences in neurovirulence that are linked to the genotype. MLVA (Multilocus Variable Number of Tandem Repeats Analysis) complex-A LM-strains replicate (Figure 4) and spread more efficiently in hippocampal slices than do complex-C ones (Balandyte et al. ,2011).
Brain slices for infection assays with scrapie prions were obtained from slaughtered lambs and caprine kids. Sets of tissue-slices were prepared from the hippocampus and the cerebellum. The slices were categorized as being either susceptible or resistant to classical scrapie, depending on the genotype of the animal as determined by the polymerase-chain reaction. Slices derived from classical scrapie-susceptible animals were microinjected with classical scrapie prions, whereas those derived from classical scrapie-resistant (and hence atypical scrapie-susceptible) animals were microinjected with atypical scrapie material. At least one slice per animal was injected with phosphate-buffered saline (negative control). Slices were then harvested at weeks 0, 3, 4, 5 and 6 post-infection. After homogenization, they were treated with Proteinase K to distinguish between PrPc and PrPsc. Protein extracts were then analyzed by PrPsc-Western blotting. Prion replication was not observed in slices that had been incoculated with atypical scrapie material. In the Western-blot analysis that was conducted at the 4-week juncture, the classical scrapie-inoculated brain-slices of two caprine kids and of one lamb revealed bands that reacted with the two prion-protein-specific antibodies. However, their molecular masses did not match with the profiles that were indicative for the disease-associated prion protein. Moreover, the signals were at the detection limit and only poorly reproducible when analyzed in replicates. It was not possible to analyze the slices of these three animals that were harvested at later time-points owing to their extensive disintegration. The other classical scrapie-inoculated brain-slices were negative in the Western-blot analyses.
Conclusions and Relevance for 3R
Although the suitability of the system for long culturing times lends itself to the investigation of slow-growing agents such as prions, we were not able to efficiently propagate scrapie prions in ovine and caprine brain-slices. However, there may be some scope for improvement by combining tissues from highly genetically-susceptible sheep or goats with scrapie strains that are specifically adapted to these genotypes.
Slaughterhouse-derived brain-slices from bovine animals were sufficiently viable and could be successfully infected with LM (Guldimann et al., 2012). Results from our infection assays with LM indicate that strains from all three genetic complexes (Balandyte et al., 2011) are capable of evoking encephalitis and that all LM-strains have to be considered as potentially neurovirulent. However, together with our MLVA-data (Balandyte et al., 2011) - according to which most encephalitic strains from ruminants cluster strongly in the genetically-homogenous complex A - our infection assays revealed the latter to be particularly neurovirulent.
Organotypic brain-slices that are derived from slaughterhouse material appear to be an adequate in-vitro model to study the pathogenesis of the intracerebral phase of LM-infection and to screen LM-strains for neurovirulence. We are confident that such organotypic brain-slice cultures represent an ethically-sustainable and host-specific in-vitro system for the investigation of neurolisteriosis. Brains from slaughtered cattle are available on a regular basis and relevant brain regions that are adequate for culturing purposes (such as the hippocampus) are easily accessible on site. Currently, we are using bovine organotypic brain-slices to study the bacterial determinants of intracerebral replication and spreading, which are crucial for an understanding of the neuropathogenetic process. The use of brain-slice cultures not only helps to spare the life of animals, but also offers the combined advantages of reflecting the organ-specific microarchitecture of the brain and of working under controllable conditions.
Our results on CNS-listeriosis might encourage other scientists to consider this in-vitro approach as an alternative to animal experiments in their own research. We are confident that in the era of next-generation sequencing (NGS) – in which novel neuroinfectious agents will be identified - this system has the potential to substitute animal models of neuroinfectious diseases with an impact on veterinary and public health.
References
Balandyté L, Brodard I, Frey J, Oevermann A, Abril C. Ruminant rhombencephalitis-associated Listeria monocytogenes alleles linked to amultilocus variable-number tandem-repeat analysis complex. Appl Environ Microbiol. 2011 Dec; 77(23):8325-35. doi:10.1128/AEM.06507-11.
Guldimann C, Lejeune B, Hofer S, Leib SL, Frey J, Zurbriggen A, Seuberlich T, Oevermann A. Ruminant organotypic brain-slice cultures as a model for the investigation of CNS listeriosis. Int J Exp Pathol. 2012 Aug;93(4):259-68. doi:10.1111/j.1365-2613.2012.00821.x.
Guldimann C, Schock A, Otter A, Frey J, Oevermann A. Listeria monocytogenes from ruminant’s abortion, enteritis and mastitis do not share common MLVA profiles with strains from ruminant’s rhombencephalitis. Submitted.
Guldimann C, Bärtschi M, Frey J, Zurbriggen A, Seuberlich T, Oevermann A. Spread and replication efficiency of Listeria monocytogenes in organotypic brain slices is related to genetic complex. In preparation.
Figures

Figure 1: Hippocampal and cerebellar brain slice culture from a bovine. Anatomical architecture is maintained and at the edges cells grow out.
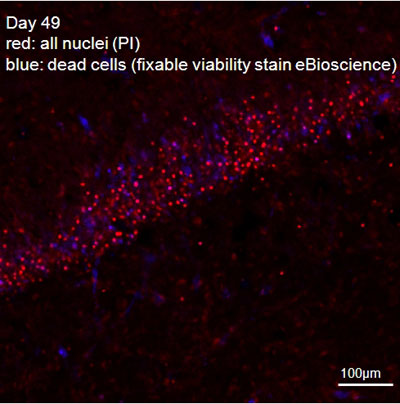
Figure 2: Hippocampal slice, combined fixable-viability (blue, nuclei of dead cells) and PI (red, all nuclei) stain.
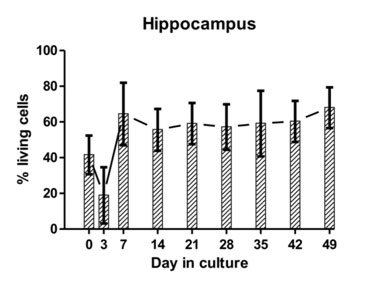
Figure 3: The average proportion of viable cells as estimated from dead cell and total cell counts are indicated at different time points for hippocampal and cerebellar tissue-slice cultures from six different calves (error bars represent standard deviations).
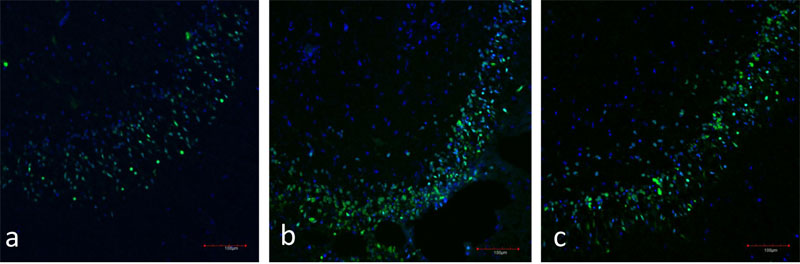
Figure 4: a) Immunofluorescence of the hippocampal dentate gyrus (day 7 in vitro). Neurons are stained in green (NeuN), nuclei are stained in blue (TOTO-3).
b) Immunofluorescence of the hippocampal dentate gyrus (day 28 in vitro). Neurons are stained in green (NeuN), nuclei are stained in blue (TOTO-3).
c)Immunofluorescence of the hippocampal dentate gyrus (day 49 in vitro). Neurons are stained in green (NeuN), nuclei are stained in blue (TOTO-3).
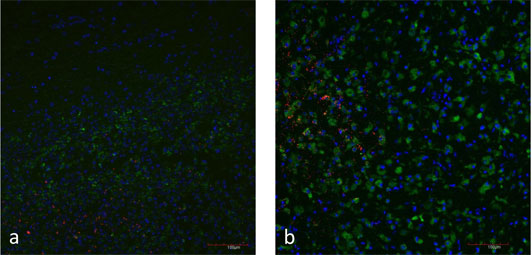
Figure 5: a) Natural case of listeric rhombencephalitis, double-immunofluorescence for L. monocytogenes (red) and microglia cells (CD68 in green). Focal replication of L. monocytogenes.
b) Brain slice inoculated with L. monocytogenes (red). Microglia cells are stained with CD68 (green). Focal replication of L. monocytogenes.
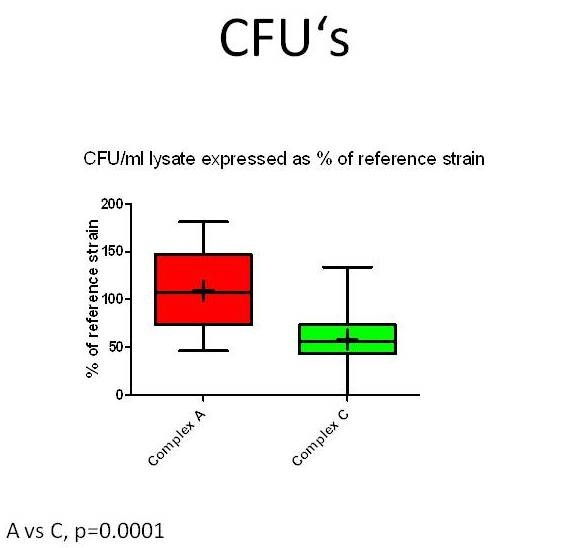
Figure 6: CFU’s recovered from lysed hippocampal slices inoculated with Listeria monocytogenesstrains (n=47) from MLVA complexes A and C. Values are expressed as % of an internal reference strain (L104). Complex A strains have significantly increased CFU’s compared to complex C strains indicating they replicate more efficiently in hippocampal slices.
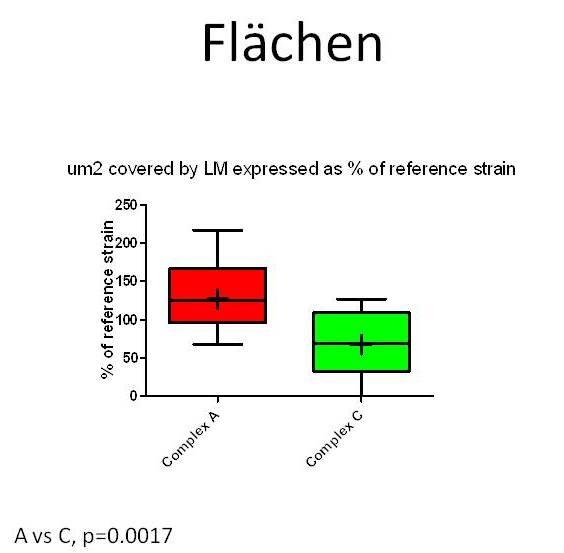
Figure 7: µm2 area of hippocampal slices invaded by Listeria monocytogenes strains from MLVA complexes A and C. Values are expressed as % of an internal reference strain (L104). Areas occupied by complex A strains are significantly larger than areas occupied by complex C strains indicating complex A strains spread more efficiently between cells in hippocampal slices.