
3R-INFO-BULLETIN 54
| | March 2015Author
Dr. Christian Heinis is an assistant professor in the Institute of Chemical Sciences and Engineering at the Ecole Polytechnique Fédérale de Lausanne (EPFL). His Laboratory of Therapeutic Proteins and Peptides (LPPT) has developed an efficient method for the generation of peptide ligands, which bind to protein targets in an antibody-like manner. The method is based on phage display and does not involve animal experimentation. Christian Heinis’s laboratory has also generated an antibody phage-display library, which is provided free of charge and without licensing constraints to any interested laboratory. Address: Prof. Christian Heinis
christian.heinis@epfl.ch Institute of Chemical Sciences and Engineering, Ecole Polytechnique Fédérale de Lausanne (EPFL)
CH-1015 Lausanne
Switzerland
Editor
Ernst B. Hunziker, Scientific Adviser of the 3R Research Foundation |
New tools for the generation of peptide- and antibody-based ligands in vitro: Reduction of antibody production in experimental animals
Antibodies are widely used as therapeutics and research reagents. For the latter application, numerous animal experiments are yearly performed to produce antibodies by immunization. Antibodies can in principle be generated also using powerful in-vitro techniques, such as phage display, which do not involve animal experimentation. However, the complex technical procedures and the limited access to phage-display libraries have hindered a wider application of in-vitro techniques for antibody development. In project No. 131-12, the research group of Christian Heinis has developed a simple and efficient phage-display strategy for the generation of affinity reagents that are based on bicyclic peptides. The bicyclic peptides bind via their two peptide rings to molecular targets in a manner that resembles the interaction of antibodies – via the complementarity-determining regions of their surface loops – with antigens. Christian Heinis’s group has also generated a large antibody phage-display library, which is provided to interested users free of charge and without any licensing constraints. This facility should promote a wider application of the phage-display technique for the generation of antibodies.
Simple and efficient methodology for the development of peptide ligands
Many research techniques, including fluorescence microscopy, Western blotting and FACS, depend upon the use of affinity reagents that bind tightly and with high specificity to proteins of interest. Currently, these affinity reagents are produced mainly by the immunization of experimental animals.
Christian Heinis’s laboratory is specialized in the development of bicyclic peptide ligands by the phage-display technique. The bicyclic peptides contain two peptide loops, which can interact with protein targets in a manner that is akin to the binding of antibodies – via the complementarity-determining regions of their surface loops – to antigens. Hence, they be seen as ‘mini-antibodies’ (Figure 1). They bind with dissociation constants in the nanomolar or even picomolar range. It has been shown that they can be applied instead of antibodies as research tools. Moreover, the small molecular size of the bicyclic peptides has rendered their commercial production by chemical synthesis inexpensive.
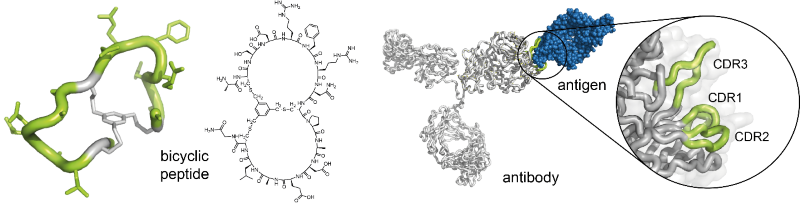
Fig 1.: Bicyclic peptides as ‘mini-antibodies’. Bicyclic peptides can be seen as miniaturized antibodies (left). They can bind to target proteins via their two peptide rings in a manner that resembles the interaction of antibodies with antigens via the complementarity-determining regions (CDRs) (right)
In order to facilitate the generation of such ‘mini-antibodies’ by the phage-display technique, Christian Heinis’s laboratory has developed a simple and efficient strategy (Figure 2), which can be applied in non-specialized laboratories.
The provided library, which is comprised of several billions of bicyclic-peptide phages, is run once through an affinity column bearing an immobilized target protein. The DNA of the captured phage is sequenced by high-throughput sequencing (using a service provider). The around 105-106 sequenced peptides are analyzed by a software, which was developed in Christian Heinis’s laboratory to identify peptides sharing consensus sequences. Peptides are then chemically synthesized (by a service provider). Unlike other phage-display strategies, the new procedure omits iterative cycles of affinity panning and phage propagation, which renders it a simpler and a more rapid technique. The novel strategy was published in Nucleic Acids Research.[1]
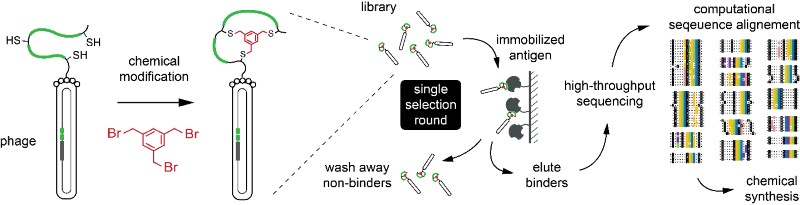
Fig 2.: Phage selection of bicyclic peptides in a single round of affinity panning. Phage-selection procedure. Peptides on the phage are chemically cyclized and subjected to an affinity selection. After a single round of selection, more than 100’000 isolated peptides are identified by sequencing the phage DNA using a high-throughput technique (left). Specific binders are identified by comparing consensus motifs (right).
Antibody phage-display library
For the development of antibodies using in-vitro techniques, such as phage display, high-quality libraries are required. Several academic groups have generated antibody phage-display libraries which they have made available to other research laboratories. However, most of the published libraries are not kept as stocks and are no longer distributed. Antibody phage-display libraries that are offered by companies are typically very expensive or available only with licensing constraints. In order to offer better access to antibody phage- display libraries, Christian Heinis’s laboratory has cloned a large scFv-antibody phage-display library, which is provided free of charge and without any licensing conditions to interested laboratories.
The antibody library was designed along the lines of others that have been reported by specialized laboratories, such as those of Prof. Dario Neri (ETH, Zurich) and Dr. Nicolas Fischer (Novimmune, Geneva). The scFv-library is based on a human antibody framework. The amino-acid sequences of the two CDR3s in each of the two variable domains are modulated (Figure 3). The library is comprised of 1.38x109 antibody clones and is stored as a stock of phage-infected bacteria in glycerol (Figure 3).
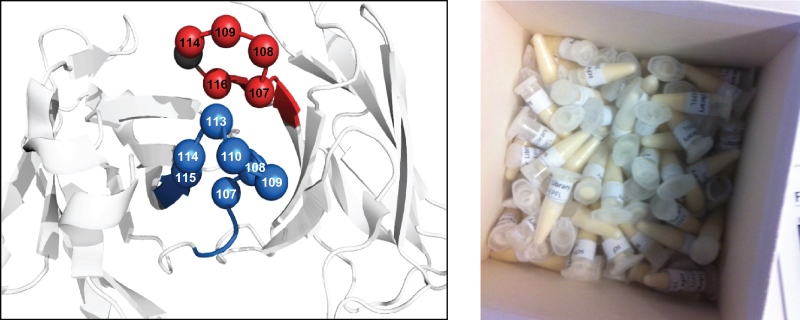
Fig 3.: Antibody phage-display library. Design of an scFv-antibody phage-display library (left). The library is based on a
human framework. The two CDR3s are randomized as indicated. Glycerol stocks of the library are provided free of charge (right).
PDF version of this Bulletin No. 54
References:
- Rebollo, I. R., Sabisz, M., Baeriswyl, V., and Heinis, C. (2014) Identification of target-binding peptide motifs by high-throughput sequencing of phage-selected peptides, Nucleic Acids Research (in print).




