 |
de | fr | en print view![]()
3R-Project 131-12
Antibody-phage-selection strategy for application in non-specialized laboratories
Michal Sabisz and Christian Heinis
Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland
christian.heinis@epfl.ch
Keywords: human; phage display; reduction; replacement
Duration: 2 years Project Completion: 2014
Background and Aim
Antibodies are widely used in research for the detection or purification of proteins. Although antibodies with tailored binding specificities can be generated by in-vitro methodologies such as the phage-display technique (1, 2), most of the antibodies that are used in research are still produced by the immunization of animals. For technical and economical reasons, in-vitro techniques are not broadly applied to generate monoclonal antibodies. Phage-display involves many experimental and error-prone steps. Moreover, it is costly to establish the complex method in new laboratories. Another limitation is that many of the existing antibody–phage-display libraries have intellectual-property constraints.
Method and Results
We have cloned a large scFv-antibody phage-display library that is available to users free of charge and without intellectual-property constraints. The library was designed to contain VH- and VL-antibody domains, in the orientation VH-linker-VL, which are connected by a flexible (GGGGS)3-linker. As an acceptor framework, human VH 3-23- and VL kappa 1-33-germline genes were chosen. 4-10 amino acids were randomized in VH CDR3 and 4-7 in VL CDR3. The library is comprised of 1.4 billion clones.
In addition to generating the scFv-phage library, we developed a phage-display-selection strategy that involves significantly fewer experimental steps than is usual, and which should facilitate the in-vitro generation of affinity ligands by non-experts. In brief, a phage-display library is subjected to a single round of affinity panning. The DNA of the isolated clones is sequenced using a next-generation sequencing technology, which companies now provide at an affordable price. We applied the approach for a peptide-based phage-selection of ligands. The procedure has been published in Nucleic Acid Research (4).
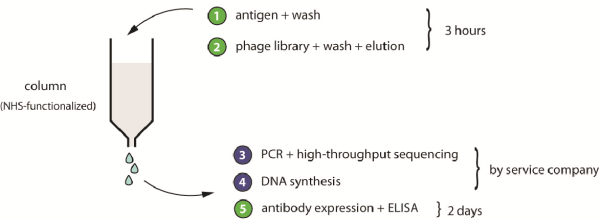
Figure 1: Schematic representation of the proposed phage-selection approach. Steps with green numbers are performed in-house by research laboratories; steps in blue are outsourced to service companies.
Conclusions and Relevance for 3R
The proposed methodology should replace animal experiments that are commonly performed to develop polyclonal and monoclonal antibodies. Monoclonal antibodies are typically produced by the repetitive injection of antigens into mice or other animals. The scFv-phage-display library has already been applied in several laboratories - including our own - to generate monoclonal antibodies. The antibodies were generated without having recourse to the immunization of animals. The novel methodology, which combines in-vitro evolution with high-throughput sequencing, is technically less complex than the classical phage-display technique. It should improve the accessibility of phage-display to non-experts.
References
(1) Rentero, I., and Heinis, C. (2011). Screening of large molecule diversities by phage display. Chimia (Aarau) 65, 843-845.
(2) Lee, C.M.Y., Iorno, N., Sierro, F. and Christ, D. (2007). Selection of human antibody fragments by phage display. Nature Protocols, 2, 3001 – 3008.
(3) Heinis, C., Rutherford, T., Freund, S., and Winter, G. (2009). Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol. 5, 502-507.
(4) Identification of target-binding peptide motifs by high-throughput sequencing of phage-selected peptides. Rentero Rebollo, I., Sabisz, M., Baeriswyl V. and Heinis, C. Nucleic Acid Research (2014), in press.
| Last modified 2018/10/12 |